Contact Admission
International Collaboration
Microbiological agents causing community pneumonia require hospitalization
P.H.Van1, 7 *, N.V.Th Thành2, T.V.Ngoc3, N.Đ.Duy4, L.T.T.Huong5, C.T.M.Thuy6, L.K.Van3
1 Laboratory of Nam Khoa company, 2Vietnam's tuberculosis and lung disease, 3Bothinology BV. Cho Ray, Respiratory Flower BV. Pham Ngoc Thach,
5 Respiratory Respiratory BV. Gia Dinh People, 6 Respiratory Respiratory BV. Can Tho Central General Department, 7 Phan Chau Trinh University, * Main responsibility
REAL RESEARCH RESULTS 2016-2017
Summary
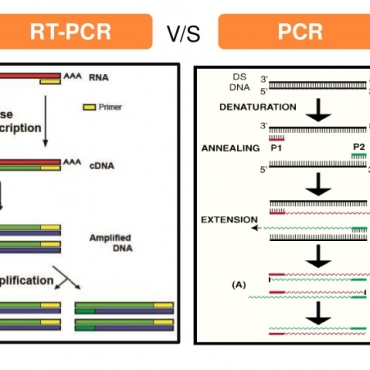
Overview: Due to the current difficult to overcome limitations in the technique of culturing sputum samples, there are almost no studies in Vietnam that show that the spectrum of microbiological agents causing community pneumonia requires hospitalization. . That is why the application of multiplex real-time PCR technique is essential to help break this deadlock situation.
Objective: Using both traditional microbiological culture methods and multiplex real-time PCR method to detect microbiological agents causing pneumonia in hospitalized community, thereby analyzing the obtained results. .
Objects and methods: This is a multicentre study conducted on community pneumonia patients with or without COPD to be hospitalized. The research method is to use the traditional microbiological method and multiplex real-time PCR method to detect the pathogens present in the sputum samples or the bronchial gas wash from the patient immediately after delivery. into research.
Results: In all, 145 patients with community pneumonia and 126 patients with COPD exacerbation infection were enrolled in the study. The multiplex real-time PCR results showed that in 69% of cases, the probiotic agents with S. pneumoniae and H. influenzae were detected with the highest rates (41.3% and 22.2%). These are K. pneumoniae (11.4%), A. baumannii (10.7%), E. coli (6.6%) and P. aeruginosa (6.3%), in addition, there are other agents detected at a lower rate. . In the culture results, excluding the isolates of Viridans streptococci, the detection rate of pathogens is 45.4%, lower than the multiplex real-time PCR method, of which the highest rate is K. pneumoniae ( 12.2%), P. aeruginosa (9.6%), A. baumannii (7.6%) and E. coli (5.2%); in no case, S. pneumoniae could be isolated and only 1.1% could isolated H. influenzae.
Discussion: If only based on the microbiological culture method, the community bacteria such as S. pneumoniae and H. influenzae will not play a role in causing pneumonia in the community and so it is very contradictory with the information. news from classic documents. Therefore, the real-time PCR solution has actually shown the microbial spectrum that actually causes community pneumonia because the results are not different from the classic studies.
Conclusion: In order to detect the microbiological agent that causes the community pneumonia, the application of real-time PCR technique is really necessary and this solution is now very feasible in both technical and economic terms. health.
Keywords: Microbiological agent causing community pneumonia; Real-time PCR detects community pneumonia
Thanks
We would like to thank Nam Khoa company for sponsoring this research.
Thank you to the young scientists of service department 2 of Nam Khoa Company
overview
Lower respiratory tract infections pose a medical burden, especially in hospitalized patients as it is difficult for doctors not to overuse strong antibiotics for treatment as the majority of microbiological results return microbiological results. Hospital bacteria [1,2] that are seldom community bacteria. The reason is that the majority of patients have used antibiotics before, so the pathogenic bacteria, although still alive in the epithelial lining of the alveoli, in the sputum sample is the main specimen for investigation, the bacteria This is dead. Not only that, the subjective reasons of the laboratory also reduce the ability to cultivate pathogenic bacteria, such as: (1) the microbiological laboratory does not have the appropriate media for isolation. the main pathogenic bacteria, which are inherently common but are difficult to cultivate; (2) the patient sample is not inoculated immediately to increase the chance of culturing pathogenic bacteria; (3) failure to assess the reliability of phlegm samples for removal of non-phlegmous samples but of oropharyngeal mucus; and (4) finally do not know how to select the pathogenic bacteria growing on the isolate agar. Therefore, to be able to reveal the true face of the spectrum of microbiological agents causing lower respiratory infections to be hospitalized, in addition to traditional microbiological testing, culture, it is necessary to use techniques. modern microbiology, which is real-time PCR technique, because it is a technique with very high sensitivity and specificity no different from culture [3].
Objectives of the study
The aim of the study is to use traditional microbiological culture techniques in combination with real-time PCR to detect pathogens present in reliable sputum samples taken from infected patients. acute lower respiratory tract (community pneumonia) must be hospitalized for treatment.
Subjects and research methods
This is a prospective study that describes cross-sectional subjects that patients with acute lower respiratory tract infections are hospitalized for treatment at the respiratory department of Cho Ray Hospital (CR), Department of Respiratory Department of Pham Hospital. Ngoc Thach (PNT), respiratory department of Gia Dinh People's Hospital (NDD), and respiratory department at Can Tho Central General Hospital (CT). The selected hospitals were those with microbiological laboratories capable of and standardized procedures for the investigation and culture of sputum or sputum samples. The study period is from May 2016 to the end of December 2016. Admission includes patients with a diagnosis of community pneumonia (CAP) who must be hospitalized and patients with acute COPD infection (CAP / COPD) who must be hospitalized. Criteria for determining community pneumonia patients hospitalized were [4,5]: (i) had systemic infections such as fever, fatigue, and anorexia; (ii) respiratory muscle symptoms such as cough, chest heaviness, sputum production, difficulty breathing; (iii) physical symptoms on lung examination such as bursts, bronchial breathing, pulmonary congestion syndrome, triple syndrome; (iv) the above conditions appear acute in 4-10 days with no explanation for any other condition; (v) Chest radiograph with new images of infiltrates that are appropriate or at least unexplained for any other previously known condition such as cancer, tuberculosis; (vi) CRP> 50mg / L. Criteria for a diagnosis of COPD when there is at least one of the following two conditions: (i) has been diagnosed and managed at a respiratory specialist facility with lung function measurement; (ii) over 40 years of age, smokers 20 pack-years or more, have chronic respiratory symptoms (cough, difficulty breathing, coughing up sputum) and / or have frequent inhaled bronchodilators; (ii) is accompanied by one of the two criteria above, which is a chest radiograph without any other abnormalities that could be the cause of chronic respiratory symptoms. Criterion for the diagnosis of an exacerbation of infection in a COPD patient in this study was (i) had 3 new symptoms including increased dyspnea, increased sputum production and increased mucus, or had the 2 above symptoms but one of which is increased purulent [6]; (ii) CRP> 15 mg / L [7]. Patients with a diagnosis of nosocomial pneumonia, pulmonary tuberculosis or at least another cause other than pneumonia producing infiltrates were not included on the chest radiograph. In addition, patients with non-qualified sputum samples or specimens containing sputum were excluded from the study.
The patient examined in this study was sputum obtained immediately after the patient was diagnosed with CAP or CAP / COPD for admission to the study and before giving the patient antibiotics. To remove the sputum, the patient is asked to rinse the mouth with sterile physiological saline before spitting out the sputum. Sputum is placed in a sterile vial with a tight cap. In case the patient is unable to cough up sputum, the specimen is the fluid drawn from the trachea taken through the nose or endoscopy. After the specimens are collected, they are immediately transferred to the hospital microbiological laboratory. In the microbiological laboratory, the sample was made a Gram stain and observed under the x100 optical field. The sample is considered reliable for culture in the presence of fields with> 25 neutrophils / field cells and <10 epithelial cells / field. The reliable samples are performed quantitatively or semi-quantitatively according to the routine laboratory used. After incubation, the samples were kept in the freezer at least -18 ° C and transferred to the laboratory of Nam Khoa Company in ice-gel containers to be conducted real-time PCR assay to detect pathogenic microorganisms in the sample.
The real-time PCR process for detecting microbiological agents present in the sputum samples or sputum specimens at Nam Khoa's laboratory is summarized as follows: (1) First of all, the sample is homogenized by one volume of sputum-homogeneous solution is PBS containing NALC (N-Acetyl L-Cysteine). (2) The sample was then extracted nucleic acid on the Thermo KingFisher FLEX automatic extraction machine with the NKRNADNAprep-MAGBEAD reagent set of Nam Khoa company, which was verified by comparison with the method. BOOM nucleic acid extraction method, RNA extraction Trizol-LS method and Roche MagnaPure nucleic acid extraction method [8]. (3) Nucleic acid extracts from phlegm samples are subjected to real-time PCR using specific primers and taqman Probes to detect and quantify microbiological pathogens including: (i) bacteria in the community are S. pneumoniae, H. influenzae, H. influenzae type b, M. catarrhalis, group A Streptococci, group B Streptococci, and S. suis; (ii) atypical bacteria are M. pneumoniae, C. pneumoniae, B. pertussis, B. parapertussis, and L. pneumophila; (iii) the hospital bacteria were methicillin-resistant or sensitive S. aureus, methicillin-resistant or sensitive S. aureus, Pantom-Valentine Leukocydine, E. faecalis, E. faecium, E. coli, K. pneumoniae, P. aeruginosa, and A.
Result
There were 145 patients with community pneumonia (21 from CR, 73 from CT, 37 NDGD, 14 from PNT) and 126 patients with exacerbation of COPD (52 from CR, 30 from CT , 16 words of physicians, 28 words of women) were included in the study. Table 1 below presents the number of cases as well as the proportion of major pathogens and co-agents present in the sputum or sputum samples detected by multiplex real-time PCR.
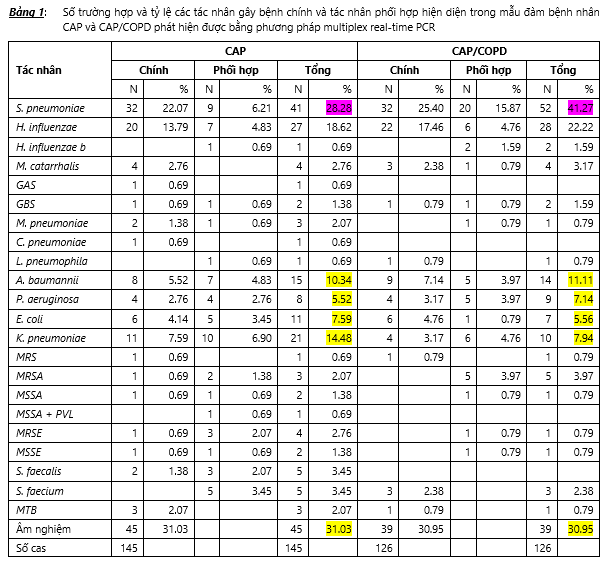
Comparing the detection rate of the pathogen on the sputum samples of the CAP and CAP / COPD groups, we found that the detection rate of the pathogen was 68.97% (100/145) in CAP patients. and 69.05% (87/126) in CAP / COPD patients and the difference was not statistically significant. The highest detectable microbiological agent was S. pneumoniae with a rate of 28.28% in CAP patients and 41.27% in CAP / COPD patients and this difference was statistically significant at the P <probability threshold. 0.05, the main reason is that 15.87% of S. pneumoniae acts as a co-agent in CAP / COPD patients, while in the CAP group it is only 6.21%. The microbiological agent with a lower incidence was H. influenzae with 18.62% in CAP patients and 22.22% in CAP / COPD patients and the difference in the rates in the two groups was not statistically significant (P> 0.05). The detection rate of community bacteria such as M. catarrhalis, atypical bacteria in CAP and CAP / COPD groups is quite low. Among the hospital bacteria, the most significant were the detection rate of A. baumannii 10.34% in the CAP group and 11.11% in the CAP / COPD group, the detection rate of K. pneumoniae in the CAP and CAP / COPD groups. is 14.48% and 7.94% and the difference in the detection rate of hospital microbial agents in the CAP and CAP / COPD groups was not statistically significant. In the CAP group, the single agent detection rate was 41.38% (60/145) and the detection rate for the multi agent was 27.59% (40/145); in the CAP / COPD group, the rate of single agent was 38.89% (49/126) and multi agent was 30.16% (38/126). There was no statistically significant difference in the rate of the single agent between the two groups and the rate of multiple agents between the two groups. Through the above results and analysis, it can be seen that the spectrum of pathogens detected by real-time PCR technique in CAP and CAP / COPD groups is not different. The spectrum of pathogens detected by multiplex real-time PCR, after pooling both groups, is presented in Table 2.
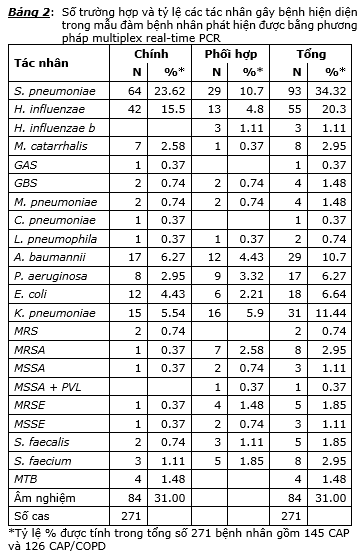
The culture results showed that the culture rate [+] in the CAP patients was 64.14% (93/145) and the CAP / COPD was 49.21% (62/126) and the difference in the culture rate [+] between the two groups was statistically significant (P <0.05). The main reason for this difference is that there has been a relatively high proportion of Streptococci bacteria belonging to the viridans group, including Streptococcus mitis and Streptococcus parasanguinis, which have been cultured oropharynx in culture. The result is the causative agent (23 cases in CAP and 9 cases in CAP / COPD). If you place these results in the culture group [-] then the actual culture rate [+] in CAP is 48.28% (70/145) and in CAP / COPD is 42.06% (53/126) and the difference These two rates are not statistically significant. Analyzing the results of culture in CAP group, the highest rates were K. pneumoniae 15.86% (23/145) and viridans streptococci 15.86% (23/145), followed by A. baumannii 7.59% (11/145) and P aeruginosa 7.59% (11/145), lower than E. coli 6.90% (10/145), yeast 4.14% (6/145). Other bacteria such as other Gram [-] bacilli (GNR), E. cloacae, E. aerogenes, S. aureus, S. epidermidis, M. catarrhalis, and H. parainfluenzae accounted for a very low percentage of 2% or more. down (ie each type only implants [+] from 1 to 3 cases). Analyzing the culture results in the CAP / COPD group, the highest [+] implantation rate was P. aeruginosa 11.90% (15/126), followed by A. baumannii 7.94% (10/126) and K. pneumoniae 7.94% (10/126), then viridans streptococci 7.14% (9/126), H. parainfluenzae 4.76% (6/126), other Gram [-] bacilli 3.97% (5/126), E. coli 3.17% (4/126), H. influenzae 2.38% (3/126). 1 case of E. aerogenes culture and 1 case of yeast culture. In both CAP and CAP / COPD groups, there was no case of S. pneumoniae implantation, while the rate of [+] culture of H. influenzae as well as M. catarrhalis was very low. Considering the agents with high implantation rate [+] such as K. pneumoniae, A. baumannii, and P. aeruginosa, the implantation rate [+] in both CAP and CAP / COPD groups is no significant difference. statistical meaning. Table 3 details the number of cases and implantation rates [+] in the CAP and CAP / COPD groups. Through the obtained culture results, it can be seen that the true culture rate [+] is low and can only detect the nematodes of hospital bacteria such as: A. baumannii, K. pneumoniae and P. aeruginosa while the culturing rate [+] of the community bacteria such as S. pneumoniae, H. influenzae and M. catarrhalis was very low or even undetectable like the case of S. pneumoniae. Not only that, culture can also give erroneous results when answering culture results [+] bacteria which are inherently pharyngeal contaminants like Viridans streptococci.
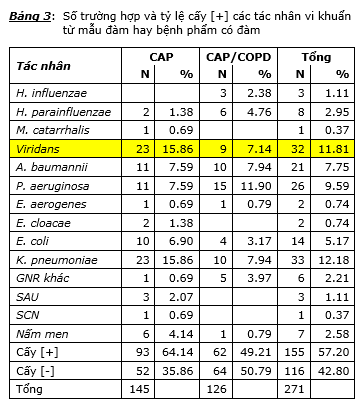
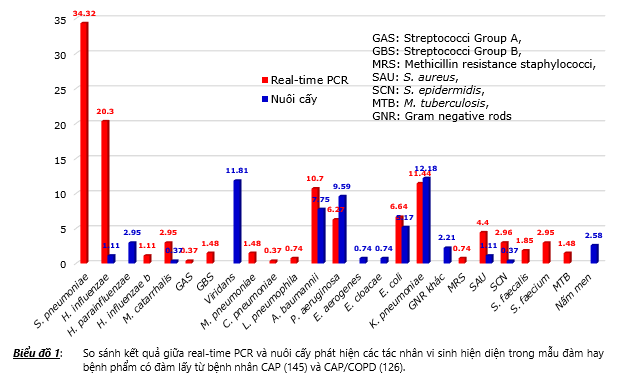
The multiplex real-time results and the culture results both showed that the detection rate of the pathogens in the CAP and CAP / COPD groups was not statistically significant. Therefore, to compare real-time PCR results with culture results, it is not necessary to analyze the two CAP groups with CAP / COPD alone, but to analyze all 271 pooled patients of both CAP and CAP / COPD groups. The results presented in chart 1 showed that real-time PCR showed the highest detection rate of community bacteria including S. pneumoniae 34.32%, followed by H. influenzae 20.3%. Meanwhile, S. pneumoniae was not detected in culturing, while the culturing rate [+] H. influenzae was also very low 1.11%. The re-cultured results showed that the percentage of hospital bacteria cultured [+] was the highest, which were the agents K. pneumoniae 12.18%, P. aeruginosa 9.59%, A. baumannii 7.75%, and E. coli 5.17%.
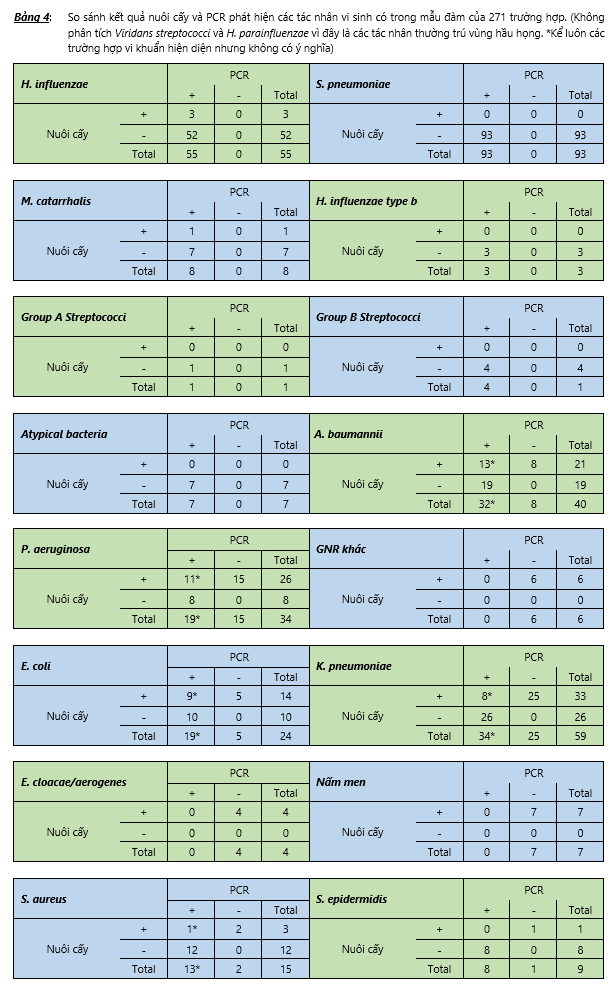
The results presented in Table 4 show that there are agents that cultured positive but not detected in the real-time PCR results, the reason is that the multiplex real-time PCR in this study has not included Specific primers and taqman probe to detect these agents. These are agents H. parainfluenzae, viridans streptococci, E. aerogenes, E. cloacae, yeast, other GNRs that are not intestinal bacteria nor are A. baumannii or P. aeruginosa.
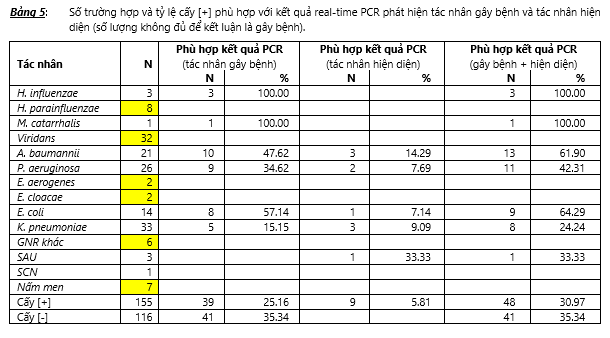
For agents that multiplex real-time PCR introduced primers and taqman Probes to detect, the cultured results still have deviations from the PCR results. Table 5 shows the number of cases (N) and the rate (%) of [+] implantation [+] consistent with the real-time PCR results in which there are cases of PCR detected as the causative agent, but there are also cases. Detectable PCR without pathogenicity (presence agent). There were 3 cases of [+] H. influenzae and 1 case of [+] M. catarrhalis culture, all of these 4 cases were detected by multiplex real-time PCR as the causative agent. Analyzing in detail cases of mismatch between transplant results [+] and multiplex results real-time PCR, we see: (1) In 21 cases of transplanting [+] A. baumannii, there were 10 cases of multiplex real-time PCR was detected as causative agent, 3 cases of real-time PCR detected only as agent present but not as pathogen due to quantity lower than 105 copies / ml. Thus, there were 8 cases of multiplex real-time PCR results undetectable, of which 7 cases were samples from patients of Respiratory Department of Can Tho Central General Hospital (CT) and 1 was from the disease. of the Respiratory Department of Gia Dinh People's Hospital (family member). The appropriate ratio between the results of culture [+] A. baumannii and multiplex real-time results on the samples at these two hospitals was 12.50% (1/8) for CT and 66.67% (2/3) for with NDD family. (2) In 26 cases of [+] P. aeruginosa culture, 9 cases of multiplex real-time PCR detected as the causative agent and 2 cases of real-time PCR detected as the agent present only. Thus, there were 15 cases of multiplex real-time PCR results undetectable, of which 10 were samples from CT and 5 were from patients of the Hospital Respiratory Department Pham Ngoc Thach (PNT). The matching rate between the results of culture [+] P. aeruginosa and the multiplex real-time results on the samples at these two hospitals was 16.67% (2/12) for CT and 50% (5/10) for with PNT. (3) In 14 cases of [+] E. coli culture, 8 cases of multiplex real-time PCR detected as the causative agent and 1 case of real-time PCR detected as the agent present only. Thus, there were 5 cases where the result of multiplex real-time PCR was not detected and all 5 were samples from CT. The matching rate between the results of culture [+] E. coli and the multiplex real-time results on the samples at this hospital was 37.50% (3/8). (4) In 33 cases of [+] K. pneumoniae, 5 cases of multiplex real-time PCR detected as pathogen and 3 cases of real-time PCR detected as agent present only. Thus, there were 25 cases of multiplex real-time PCR results undetectable, of which 21 were samples from CT, 2 were from NDT, and 2 were from PNT. The appropriate ratio between the results of culture [+] K. pneumoniae and the multiplex real-time results on the samples at these three hospitals was 16% (4/25) for CT, 60% (3/5) for for traders and 33.33% (1/3) for women. (5) In 3 cases of [+] S. aureus culture, only 1 case of multiplex real-time PCR detected, but only present agent, not pathogen. Thus, there were 2 cases of undetectable multiplex real-time PCR in which 1 was sample from CT and 1 from PNT. The appropriate ratio between the results of culture [+] S. aureus and multiplex real-time PCR results is 0.00% (0/1) for CT and 50.00% (1/2) for PNT.
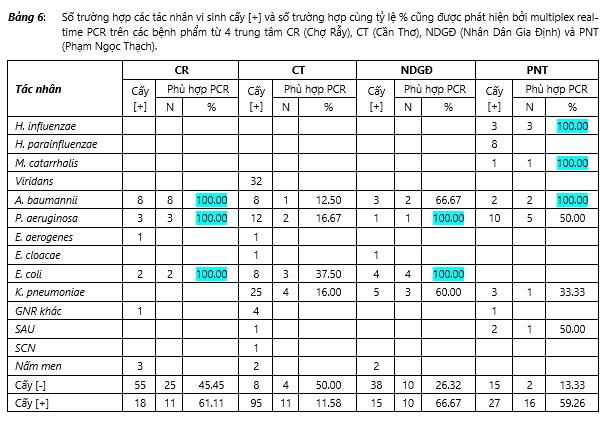
In 155 cases of [+] culture, 39 cases were detected by multiplex real-time PCR as the causative agent and 9 cases of real-time PCR detected only as the agent present. The matching ratio between the results of culture [+] and results detected by PCR is 30.97% (48/155), of which the highest rate is 66.67% (10/15), followed by CR 61.11% (11 / 18), then PNT was 59.26%, while CT was the lowest 11.58%. There are 116 cases of culture [-] in which 41 cases are the result of multiplex real-time PCR with no detectable agent. Thus, the rate consistent with PCR results in cases of culture [-] is 35.34%, in which the highest ratio is CT 50.00% (4/8), CR 45.45% (25/55), then NDF 26.32% (10 / 38). The lowest is PNT 13.33% (2/15). Table 6 presents the number of (N) cases and the rate (%) of [+] implantation [+] consistent with the real-time PCR results on the samples sent from 4 research centers, namely CR, CT, NDF and PNT.
Discuss
According to statistics of the Ministry of Health in 2014, pneumonia is a leading fatal disease with the rate of 1.32 per 100,000 people equivalent to traumatic brain injury [12]. One of the reasons that pneumonia has a high mortality rate is that it is often difficult for a doctor to target antibiotics because microbiological test results often fail to identify micro-agents. pathogenesis. We also know that the primary pathogen for detection of pathogens is the sputum or sputum samples obtained from the patient. However, the sputum test has many challenges to overcome because this is an inherently adulterated specimen due to the need to pass the oropharynx, so culturing the correct pathogen, not the contaminant bacteria. This is a huge challenge. In addition, the most common bacterial pathogens that cause lower respiratory disease are very difficult bacteria that not only require sufficient isolation media, but also require immediate inoculation of samples, and This basic requirement is often less met in clinical microbiology laboratories in hospitals. In addition to the sputum, another type of specimen that is also essential to the order to detect the bacterial agent that causes pneumonia is blood culture. However, the main challenge of a blood transplant is that the rate of blood transplants [+] in a diagnosis of pneumonia is usually below 14% because not all pathogens are capable of invading the blood, besides the results of culture. Blood is also sometimes false positives due to adulteration due to technical errors in blood cultures in bed as well as the process of blood culture monitoring in the laboratory. Serum testing for pathogen-specific antibodies is the solution that some laboratories now use to detect pathogens that cannot be cultivated routinely such as viruses or atypical bacteria. However, the test for specific antibodies of the IgG class is often not useful because it requires a double serum, while the test for specific antibodies of the IgM class has problems of sensitivity and specificity. and requires a cut-off depending on the epidemiological area. Immunochemical assays that detect antigens that dissolve S. pneumoniae and Legionella bacteria in urine are also a solution for detecting these two agents. However, due to its high cost and low sensitivity, it is rarely used. For atypical viral or bacterial agents, the ELISA technique or direct fluorescent antibody staining is also used, but these tests are also difficult to apply because the sensitivity of the majority is not high.
Based on the principle of both replicating and detecting specific nucleic acid sequences (DNA or RNA) in the sample, real-time PCR is currently considered the technique with the highest sensitivity and highest specificity in detection. The pathogenic microbiological agents are present in different specimens [3,13,14]. Recently, more and more reports have shown that real-time PCR is the most sensitive and specific solution in detecting microbiological agents that cause pneumonia or lower respiratory infections [9-11,15,16] . In Vietnam, there was a study by Takahashi K. and his colleagues in Khanh Hoa using PCR to detect microbiological agents causing community pneumonia, but this work is really is currently in the 2009 flu epidemic, so it is difficult to reflect the true microbiological spectrum that causes CAP [17]. In particular, we have also carried out research projects proving the effectiveness of real-time PCR technology in detecting microbiological pathogens causing community pneumonia in adult and hospital patients [18 -20]. The results of the above works show that: (1) the main pathogens are the micro-emergency agents in the community, in which the leader is S. pneumoniae, in addition, agents such as H. influenzae and M. catarrhalis also accounts for a significant proportion; (2) atypical bacterial agents such as M. pneumoniae also play a very important role, possibly as a major pathogen and also a coordinating role, especially pneumonia in children; (3) The culture method is almost impossible to detect the agents S. pneumoniae and M. pneumoniae are two main factors causing pneumonia in patients.
Discuss
In this study, study subjects are hospitalized community pneumonia patients, including community pneumonia patients with COPD. Specimens investigated were bronchial air-aspirating or sputum. The method of investigation is culturing and performing multiplex real-time PCR to detect pathogenic microorganisms (except for fungi). The results obtained showed the following remarkable points: (1) The results of culture were isolated and returned cases of pharyngeal bacteria like Viridans streptococci, so if we exclude the fields. In this case, the positive culture rate is 45.4% (153/271); (2) Multiplex real-time PCR for the rate of detecting the pathogen was 69% significantly higher than the culture method 45.4%; (3) There was no difference in the prevalence of pathogens detected by culture between the two groups of patients with COPD and community pneumonia, nor was there a difference in the rate of Pathogenicity detected by real-time PCR between two groups of patients with community pneumonia and community pneumonia with COPD, so it can be said that there is no difference in the rate and spectrum of bacteria causing pneumonia. community and community pneumonia have COPD although the group with COPD can be considered as a group of public health-related pneumonia (HCAP); (4) There is a difference in the spectrum of pathogenic bacteria detected by real-time PCR compared with culture, in which the culture method for pathogenic bacteria spectrum is mainly hospital bacteria such as K. pneumoniae , P. aeruginosa, A. baumannii and E. coli, while the PCR method showed the spectrum of bacteria, mainly S. pneumoniae and H. influenzae, while hospital bacteria were still detectable but at the rate lower; (5) The culturing method could not detect S. pneumoniae and other community bacteria such as H. influenzae, M. catarrhalis, the rate of detection by culture was very low; (6) Because the survey subjects are patients with community pneumonia or community pneumonia with COPD and need to be hospitalized, there must be the role of multi-resistant bacteria like K. pneumoniae, P. aeruginosa, A. baumannii and E. coli, therefore, these bacteria occupy the second rate after the two agents S. pneumoniae and H. influenzae detected by real-time PCR, and this record is also worth the attention of doctors. treatment of concern when choosing antibiotics for the first step in treatment; (7) The detection rate of atypical bacterial agents such as M. pneumoniae, C. pneumoniae, L. pneumophila is very low, and in no case of virus agent detection is a necessary feature. is noted in this study, and this feature is probably related to the population of patients surveyed for hospitalized community pneumonia cases, and for these subjects the role of bacteria always prevailed. .
Conclude
Traditional microbiological methods of culture and isolation of pathogenic microorganisms have always been considered the gold standard in diagnosing infectious agents. However, the traditional microbiological method always faces obstacles that are difficult to overcome, especially when performing on sputum samples to detect the agent causing pneumonia. Therefore, a tool that needs to be added in phytosanitary assays is the multiplex real-time PCR technique. Implementing this tool for clinical microbiology laboratories is both technically feasible as well as cost effective. Technically feasible is because the applied method is based on STREALINE REAL-TIME PCR model with sample processing done completely automatically [8], and real-time PCR technique is preferred technique. It is integrated for clinical laboratories by eliminating the need to do additional experiments to analyze results, but only need to read the results displayed on the screen thereby avoiding the risk of contamination [3]. It is feasible in terms of cost because the equipment cost is less expensive than an automatic hematology machine, and now many hospitals' clinical laboratories already have this facility available. Not only that, in terms of efficiency versus cost, we will see this tool achieve both efficiency and cost because if based on the culture results, the treating doctor will always use antibiotics step by step. the first is broad-spectrum and strong antibiotics that are cultured by bacterial agents, which are usually only multidrug-resistant nosocomial agents. In addition, because real-time PCR results are often available early, this is a method that helps the treating doctor to tell or adjust the target antibiotic early, only after 8 hours after the sample is sent for testing. Finally, this is a comprehensive study to investigate the detection of microbiological agents causing community pneumonia to be hospitalized, the results from the multiplex real-time PCR method reflect the true spectrum of microbiological agents. pathogenicity due to its ability to detect community microbial agents, which is very low compared to culture due to the difficult to overcome limitations. The results obtained from this study can be evidence for clinicians to use in the development of an initial antibiotic regimen for treatment, to avoid overuse of broad-spectrum and strong antibiotics in the first place. , thereby limiting the development of resistance to these antibiotics.
References
-
Nguyen Thi Ngoc Bich, Tran Van Ngoc. (2007). Study on characteristics of pathogenic bacteria in lower respiratory infections and antibiotic resistance at Cho Ray Hospital. Journal of Medicine and Pharmacy Information - 2nd National Lung Disease Scientific Conference - October 2007, pp. 55-59
2.Nguyen Van Hung, Pham Hoang Yen, Tran Thi Bich Thuy (2007). Antibiotic susceptibility of gram-negative bacilli isolated from lower respiratory tract specimens at tuberculosis hospital and central lung disease. Journal of Medicine and Pharmacy Information - 2nd National Lung Disease Scientific Conference - October 2007, page 49-53.
3.Pham Hung Van. (2009). PCR and real-time PCR: The basics and common applications. Medical publishing house. p3-p33
4.W S Lim, S V Baudouin, R C George et al. (2009). British Thoracic Society, Community Acquired Pneumonia in Adults Guideline Group. Guidelines for the management of community acquired pneumonia in adults: update 2009. Thorax 2009; 64 (Suppl III): iii1 – iii55.
5.Vietnam Association of Tuberculosis and Lung Disease. (2012). Guidelines for management of non-TB lower respiratory infections. Medical Publishing House, first published 2012. Tr141-p169
6.Woodhead M, Blasi F, Ewig S, et al. (2011). Guidelines for the management of lower respiratory tract infections. Clin Microbiol Infect 2011; 17 (Suppl. 6): 1–24.
7.Peng C, Tian C, Zhang Y, Yang X, Feng Y, Fan H. (2013). C-reactive protein levels predict bacterial exacerbation in patients with chronic obstructive pulmonary disease. Am J Med Sci. 2013 Mar; 345 (3): 190-4.
8.Van Pham Hung et al. (2015). The solution for the low-income countries to establish the automatic extraction of the nucleic acid from the clinical samples. Asean Congress on Medical Biotechnology and Molecular Biosciences 2015. October 8th - 9th, 2015 at Arnoma Grand Hotel, Bangkok, Thailand. Pp25-31
9.Gadsby N. J., Templeton K. E. et al. (2015). Development of two real-time multiplex PCR assays for the detection and quantification of eight key bacterial pathogens in lower respiratory tract infections. Clin Microbiol Infect. 21: 788.e1-788.e13
10.Jan J. O., Marc J. M. B. et al. (2015). Impact of Rapid Detection of Viral and Atypical Bacterial Pathogens by Real-Time Polymerase Chain Reaction for Patients with Lower Respiratory Tract Infection. Clinical Infectious Diseases. 41: 1438–44.
11.Alicia Edin et al. (2015). Development and Laboratory Evaluation of a Real-Time PCR Assay for Detecting Viruses and Bacteria of Relevance for Community-Acquired Pneumonia. The Journal of Moleculat Diagnostics. 17 (3): 315-324
12.Ministry of Health. (2014). Medical Statistics Yearbook. p214
13.Naomi J. Gadsby et al. (2016). Comprehensive Molecular Testing in CAP. CID. 62 (April 1): 817-823
14.Margret Schuller et al. (2010). PCR for Clinical Microbiology. Springer publisher. Pp11-48
15.Chae Lim Jung et al. (2010). Clinical Evaluation of the Multiplex PCR Assay for the Detection of Bacterial Pathogens in Respiratory Specimens from Patients with Pneumonia. Korean J Clin Microbiol 13 (1): 40-45.
16.Tie-Gang Zhang et al. (2015). Detection of respiratory viral and bacterial pathogens causing pediatric community-acquired pneumonia in Beijing using real-time PCR. Chronic Diseases and Translational Medicine. 1: 110-16
17.Takahashi K., Yoshida L. M. et al. (two thousand and thirteen). The incidence and aetiology of hospitalized community-acquired pneumonia among Vietnamese adults: a prospective surveillance in Central Vietnam. BMC Infectious Diseases. 13: 296.
18.Tran Thi Thanh Vy. (2014). Determining the rate of atypical bacterial agents that cause pneumonia hospitalized at Nguyen Tri Phuong Hospital during the period from November 2013 to June 2014. Medical Master Thesis.
19.Bui Le Huu Bich Van. (2015). Community pneumonia causes that do not respond to initial antibiotic treatment in children under 5 years old at the General Department of Internal Medicine 2 Children's Hospital 1. Thesis Master of Medicine specializes in microbiology. tr52-p57
20.Tran Quang Khai. (2016). Characteristics of lobe pneumonia in children at General Internal Medicine Department 2 Children's Hospital 1. Master's thesis in pediatrics. Tr60-p67
Other research
- Overdiagnosis: When the Boundaries of Disease Become Blurred ( 08:22 - 08/07/2025 )
- Continuous Medical Knowledge Tied to Better Clinical Outcomes in Internal Medicine ( 10:56 - 19/06/2025 )
- Rapid and sensitive diagnostic procedure for multiple detection of pandemic Coronaviridae family members SARS-CoV-2, SARS-CoV, MERS-CoV and HCoV: a translational research and cooperation between the Phan Chau Trinh University in Vietnam and Universit ( 15:42 - 29/06/2020 )
- PCTU studies at ISSAR 2019 international conference - Gyeongju, Korea ( 16:17 - 04/11/2019 )
- Melioidosis in Vietnam: Recently Improved Recognition but still an Uncertain Disease Burden after Almost a Century of Reporting ( 13:29 - 27/04/2018 )
- The in-vitro activity of sitafloxacin against the most common bacterial pathogens isolated from the clinical samples ( 11:10 - 03/02/2018 )
- Evaluate the role of the microbiological molecular biology tests in the early detection of pathogens causing lower respiratory infections ( 10:52 - 03/02/2018 )
- Production and evaluation of the kit using magnetic silica coated nano-iron beads to extract the nucleic acid from different samples ( 10:16 - 03/02/2018 )
- Challenges in selecting antibiotics treatment for H. Pylori in Vietnam ( 09:12 - 03/02/2018 )
- The resistance of the gram-negative rods isolated from clinical cases in Vietnam to doripenem ( 08:46 - 03/02/2018 )